H. P. Friedrichs (AC7ZL) Homepage
My Projects
Automatic Bone Cancellation for CT Angiograms
Summary
Some months back, a local radiologist and I were discussing image processing techniques, and he shared with me a journal reference to some research that was being conducted in the Netherlands. The purpose of this research was to develop software to automate the process of bone removal in CT Angiogram images. As he thought that I would have a better chance of understanding the technical details of this software than he, he asked if I would email these researchers on his behalf and inquire about their work.
I'm not sure if the software in question was not yet fully functional, or if the people I communicated with were concerned about intellectual property issues, but I remember them being vague and not particularly communicative. I reported this back to the radiologist, and then on a fluke, suggested that I might try taking a stab at this problem.
The first challenge was to figure out how to get image data from the CT scanner. Modern medical equipment of this type communicates using a data transfer standard called DICOM. DICOM data may contain numerical data, images, sounds, electrical waveforms, or just about any kind of data you can think of. I soon discovered that it is a running joke in the industry that the DICOM "standard" is so flexible and all-inclusive that it is scarcely a standard at all. Apparently, it is not uncommon for two pieces of DICOM-compliant hardware to be completely incapable of communicating with each other!
At the time, significant portions of the DICOM standard were documented on the web, so I was able to study the internal structure of DICOM files, and compare these "standards" to the data that actually appeared in sample images. I had hoped to find a DICOM "C" library on the web that would jump start my effort to read, manipulate, and write DICOM data, but I never did find source code that did what I wanted the way I needed it done. So, I wrote it all from scratch.
The heart of the bone cancellation idea is to combine the data from two data sets, one accumulated as a "reference" scan, and one accumulated while the patient is receiving a contrasting agent. As it turns out, this simple principle is a good deal more complicated to implement than it sounds.
The first problem is that patients are likely to move from one scanning session to the next. In fact, the two data sets might conceivably have occurred on different days. Numerical cancellation of bone data is not possible until alignment errors have been accounted for. Note that such errors can occur as linear offsets in the x, y, and z planes, as well as rotational error that I'll refer to as roll, pitch, and yaw.
For the purpose of proof-of-concept, I decided to limited my initial efforts to two dimensions of linear offset, and one rotational axis, on a three dimensional data set.
As if alignment was not difficult enough, it was further complicated by the fact that the images collected while contrasting agents are in use are typically scanned with higher beam currents. The upshot of this is not unlike a photograph taken of a high-contrast object in bright light. Images tend to "bloom" which distorts their true dimensions.
I eventually developed a whole array of routines to align, normalize, "de-bloom" and cancel bone from image data sets. I fed my processed data via DICOM to a 3-D imaging station, and generated some pretty neat images. Some of them can be reached through the following hyperlinks. (Use your browser's "Back" button when you are through viewing a:
Skull 1 is both an early and poor example of my bone cancellation efforts. However, the picture is so cool looking, I had to include it here!

Skull 2 is a later image, looking down into the top of the subject's cranium. In this case, the bone cancellation is quite good. Bone remnants can be seen as fine white filaments or dotted lines encircling the subject's head. The tangled tube-like objects in the center of the image are what they appear to be--blood vessels.
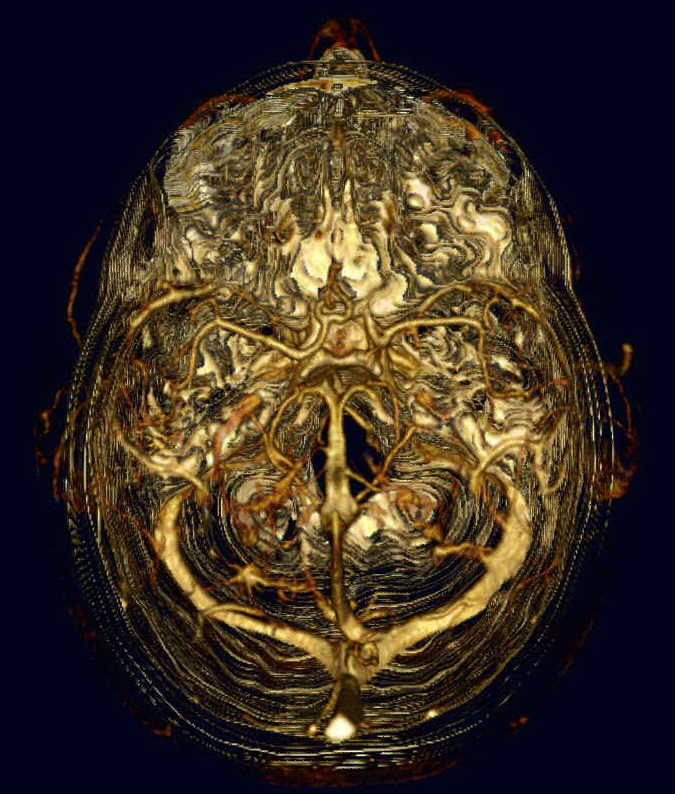
Vessel is a close-up of some of the blood vessels taken from a later set of tests. Notice that the cancellation of surrounding bone is nearly complete.
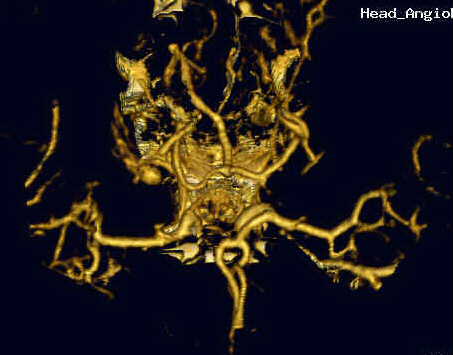
After some 9,000 lines of "C" code and several months worth of effort, I ceased further development. I had other ideas to improve the process, but I only have so much free time available, and I already had more projects than free time. This is a project I wouldn't mind revisiting someday, however. Anyone care to fund my research? <grin>
(revised 08/04/06)
(revised 04/20/09)


